

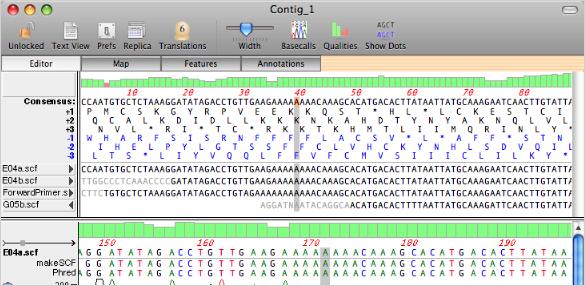
Unlike many companies in this field, we are continuing work on improving the productivity and utility of Sanger sequencing based on feedback from our collaborators around the world, and focusing on smoothly integrating those features with the latest tools for next-generation DNA sequence alignment. If you work in a lab that supports NGS, you must already have information technology support, but our mission is to make the data and analysis of ALL sequencing data accessible and understandable to life scientists without requiring that they have a degree in computer engineering. We have continued our strategy of adding functionality to Sequencher that focuses on labs doing DNA sequencing. Working with core labs that use structured naming conventions to track data for individual clients, we developed the Assemble by Name strategy that has become such a powerful and popular tool for combining multiple sequencing projects into a single analysis run. The variance table was developed in the mid 1990's and became a key element first for forensic sequencing of mtDNA, and then for virtually all of our collaborators. Gene Codes developed the Assemble to Reference Sequence strategy that is widely used to speed up assembly and assign base-numbering systems and features to new data. Multiple electropherograms can be seen simultaneously in the trace pane in order to quickly compare mutation calls between samples.Gene Codes has long been an innovator, investing in the R&D to develop powerful features for your DNA sequence analysis.
Both the mutation table pane and the base pane display variants detected in the same positions in all three patients. The sample files were grouped by a patient identifier located in the sample file names. Group Samples by Patient, Gene, or Contigįigure 3: The Project Reviewer Report shows replicate samples for three different individuals aligning to the same reference sequence. Mutations can be edited or confirmed in the trace pane and mutation table pane. The color-coded mutation table can display variants relative to genomic, CDS, mRNA, HGVS, or forensic (SWGDAM) annotation. Annotation includes c.DNA numbering at the top of the report, regions of interest and CDS identifiers in the electropherograms, and mutation calls in the mutation table. Annotation is provided by the reference GenBank or SEQ file loaded into the project. Each pane is collapsible, allowing the user to add or remove panes and readjust window sizes.įigure 2: Overlapping amplicons are displayed in the base pane along with color-coded mutation calls.
Variants are listed in the mutation pane at the bottom of the viewer, providing a convenient way to confirm or edit mutations with the click of a mouse. The trace pane provides the sample and mutation electropherograms for each sample trace. The base pane provides color-coded variants and the sample alignment to the reference as well as its overlap to other sequence traces.

#SEQUENCHER CONTIG UPDATE#
Double-clicking on a sequence trace in the sample pane will update the base and trace panes with the appropriate sequence. Interactive Viewer Features Four Linked Data Windowsįigure 1: The Project Reviewer Report of Mutation Surveyor provides four interactive and hyperlinked views of Sanger sequencing traces in a mutation project. The viewer is highly interactive, offering linked navigation between four data windows, including links to the sample and mutation electropherogram windows, and mutation report for quick and easy confirmation of variants such as single nucleotide polymorphisms (SNPs), low-frequency variants (somatic, mosaic, heteroplasmic), and homozygous and heterozygous insertion and deletion (indel) events. Viewing and editing variants is also simplified in the Project Reviewer Report, which color codes variants and provides multiple ways to view sequence traces. The Project Reviewer Report of Mutation Surveyor provides a versatile way of grouping and viewing sample files that are aligned to the same reference, including an overlapping view of individual contigs, an overlapping view of all contigs mapping to the same GenBank file, or an overlapping view of sample files associated with a patient identifier.
#SEQUENCHER CONTIG SOFTWARE#
Contig Reference Assembly and Variant Review in Mutation Surveyorįollowing Sanger sequencing trace alignment and automatic detection of variants, Mutation Surveyor software provides a unique contig reference assembly and variant viewer which would negate the need for additional software packages such as Thermo Fisher Scientific SeqScape® Gene Code's Sequencher® or JSI Medical Systems' SEQUENCE PILOT.


 0 kommentar(er)
0 kommentar(er)
